CombiUgi virtual library generation via Google Spreadsheet
Andrew Lang has just created a service that lets anyone create a virtual library of Ugi products by entering the SMILES of the starting materials in a Google Spreadsheet.
First copy this template sheet (you must use File -> create a copy - copying and pasting cells will not work). Then publish the Google Spreadsheet under the Share tab.
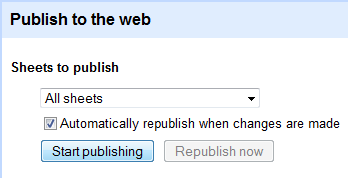
Next add the key of your new Spreadsheet (as it appears in the URL) to a URL of this form:
http://showme.physics.drexel.edu/onsc/combiugi/combiugi.php?key=tR6lhYF_iqGdmceaAg-WRLg
The resulting page, which could take a long time to load for large libraries, can then be saved as a CSV file. On Firefox this is done by selecting Save As Text File.
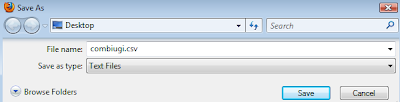 If you put a CSV extension in the name you can then open the file directly in Excel:
If you put a CSV extension in the name you can then open the file directly in Excel: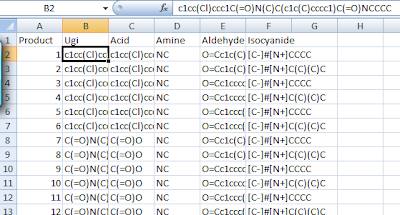
All the results are in SMILES format and using all the tricks of Excel can be sorted or filtered even by reactant. One could also copy and paste to another Google Spreadsheet to manipulate the dataset.
This service replaces the one Rajarshi Guha had set up a while back at Indiana University. A key difference with this service is that it requires SMILES to be constructed as shown in the template sheet:
- N to the left for amines
- C(=O)O to the right for carboxylic acids
- O=C to the left for aldehydes
- [C-]#[N+] to the left for isocyanides
Labels: CombiUgi, Ugi reaction, virtual library






