Dangerous Data: Lessons from my Cheminfo Retrieval Class
I'm not sure what my students expected before taking my Chemical Information Retrieval class this fall. My guess is that most just wanted to learn how to use databases to quickly find "facts". From what I can gather much of their education has consisted of teachers giving them "facts" to memorize and telling them which sources to trust.
Trust your textbook - don't trust Wikipedia.If I did my job correctly they should have learned that no sources should be trusted implicitly. Unfortunately squeezing useful information from chemistry sources is a lot of work and hopefully they learned some tools and attitudes that will prove helpful no matter how chemistry data is delivered in the future.
Trust your encylopedia - don't trust Google.
Trust papers in peer reviewed journals - don't trust websites.
I have previously discussed how trust should have no part in science. It is probably one of the most insidious factors infesting the scientific process as we currently use it.
To demonstrate this, I had students find 5 different sources for properties of chemicals of their choice. Some of the results demonstrate how difficult it can be to obtain measurements with confidence.
Here are my favorite findings from this assignment as a top 3 countdown:
#3 The density of resveratrol on 3DMET
Searching for chemical property information on Google quickly reveals the plethora of databases indexed on the internet with a broken chain of provenance. These range from academic exercises of good will to company catalogs, presumably there to sell products. Although it is usually not possible to find out the source of the information, you can sometimes infer the origin by seeing identical numbers showing up in multiple places.
But sometimes the results are downright bizarre - consider the number 1.009384166 as the density of resveratrol from what looks like a Japanese government site 3DMET. First of all no units are given but lets assume this is in g/ml. The number of significant figures is curious and suggests the results of a calculation, perhaps a prediction. In this case the source is from the MOE software. This is clearly a different algorithm from the one used by ACDLabs, which comes in at 1.356 g/ml, much more realistic when put up against all 5 sources:
#2 The melting point for DMT depends on the language
- 1.359 g/cm3 ChemSpider predicted
- 1.36 g/cm3 (20 C) Chemical Book MSDS
- 1.009384166 3DMed
- 1.41 g/cm3 (-30.15C) DOI (found with the aid of Beilstein)
- 1.359 g/cm3 LookChem
I have to admit being really surprised by this. Even though I knew that Wikipedia pages in different languages were not exact translations I would have assumed that the chemical infoboxes would not be recreated. Interestingly, the German edition has a reference but I was not able to access it since it is a commercial database. The English edition has no specific references. Here is a list of sources:
- 40–59 ºC Wikipedia English
- 47–49 ºC ChemSynthesis
- 49 ºC and 74 ºC (two different crystal structures) AllExperts Wikipedia French
- 44.6–46.8 ºC Wikipedia German
This is by far my favorite because it most clearly demonstrates the dangers of the concept of a "trusted source". From the compilation prepared by the student, this paper (Kwang08) reported the solubility of EGCG at 521.7 g/l:
 This is from a paper that spent 5 months undergoing peer review with a well respected publisher. Also it appeared recently so one would expect the benefit of the best instruments and comparison with historical values. But even beyond all of this, the numbers are in the opposite order to the point explained in the paragraph. In our system of peer review we don't expect reviewers to verify every data point - but we do expect the text to be evaluated as logically consistent.
This is from a paper that spent 5 months undergoing peer review with a well respected publisher. Also it appeared recently so one would expect the benefit of the best instruments and comparison with historical values. But even beyond all of this, the numbers are in the opposite order to the point explained in the paragraph. In our system of peer review we don't expect reviewers to verify every data point - but we do expect the text to be evaluated as logically consistent.Now if we follow the reference provided for this paragraph we find the following paper (Liang06), with this:
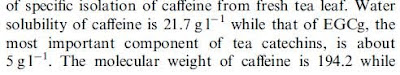 We can now see what happened: the 21.7 was accidentally duplicated from the caffeine measurement and appended to the 5 g/l for EGCG. This is a lot more reasonable, even though I am not clear about where that number comes from in this second paper.
We can now see what happened: the 21.7 was accidentally duplicated from the caffeine measurement and appended to the 5 g/l for EGCG. This is a lot more reasonable, even though I am not clear about where that number comes from in this second paper.We can get some idea of the potential source of this information from the Specification Sheet for EGCG on Sigma-Aldrich:

Notice that this does not state that the maximum solubility of EGCG in water is 5 mg/ml - just that a solution of that concentration can be made. This value is repeated elsewhere, such as this NCI document, which references Sigma-Aldrich:
 From here the situation gets muddled. Another search reveals this peer reviewed paper (Moon06), which appeared in 2006:
From here the situation gets muddled. Another search reveals this peer reviewed paper (Moon06), which appeared in 2006: Expressed in mM this translates to about 2.3 g/l. Clearly this value is inconsistent with the Sigma-Aldrich report of being able to make a clear solution at 5 g/l.
Expressed in mM this translates to about 2.3 g/l. Clearly this value is inconsistent with the Sigma-Aldrich report of being able to make a clear solution at 5 g/l.Luckily, in this case we have some details of the experiments:
 The measurements were done in triplicate and averaged. Unfortunately this does not reveal any sources of systematic error. One clue as to why these values are contradictory might be the method of dissolution. One hour sonication at room temperature might just not be enough to make a saturated solution for this compound. (Although one might expect the error to lie on the high side because the sample were diluted before being filtered) What would answer this definitively are the experimental details of how the Sigma-Aldrich source prepared the 5 g/l solution. If it went in within a few minutes without much agitation, that would be inconsistent with this hypothesis of insufficient mixing. In that case we would want to look at the HPLC traces in this paper for another type of systematic error.
The measurements were done in triplicate and averaged. Unfortunately this does not reveal any sources of systematic error. One clue as to why these values are contradictory might be the method of dissolution. One hour sonication at room temperature might just not be enough to make a saturated solution for this compound. (Although one might expect the error to lie on the high side because the sample were diluted before being filtered) What would answer this definitively are the experimental details of how the Sigma-Aldrich source prepared the 5 g/l solution. If it went in within a few minutes without much agitation, that would be inconsistent with this hypothesis of insufficient mixing. In that case we would want to look at the HPLC traces in this paper for another type of systematic error.Unfortunately, the chain of information provenance ends here. Just based on the data provided so far, there is significant uncertainty in the aqueous solubility of EGCG, similar to our uncertainty about the melting point of strychnine.
As long as scientists don't provide - and are not required to provide by publishers - the full experimental details recorded in their lab notebooks, this type of uncertainty will continue to plague science and make the communication of knowledge much more difficult than it need be.
Unfortunately the concept of "trusted sources" is being used as a building block of some major chemical information projects currently underway - WolframAlpha and the chemical infobox data of Wikipedia are prime examples. Ironically, MSDS sheets are listed as a reliable "trusted source" for the infoboxes, when they have been shown to be very unreliable (see my previous post about this with statistics). These are probably one of the most dangerous sources of information because they appear to be trustworthy - coming from chemical companies and the government - and often found on university websites. Combine that with the absence of references or experimental details and the potential for replication of errors is very high and very difficult to correct.
WolframAlpha does have a mechanism to provide information about sources but it requires submitting a reason and personal information.
 To see how this works in practice I made a request for the source of an entry with erroneous data - glatiramer acetate:
To see how this works in practice I made a request for the source of an entry with erroneous data - glatiramer acetate: I submitted this 10 days ago and still don't know the source.
I submitted this 10 days ago and still don't know the source.Rapid access to specific sources is important for maximizing the usefulness of databases. Without that it becomes very difficult to assess the meaning of reported measurements and compare with results from other databases.
It is not possible to remove all errors from scientific publication. But that's only a problem when it is difficult to determine that there are errors in the first place because insufficient information is provided.
Scientists can handle ambiguity. If you look at the discussion over the blogosphere concerning the JACS NaH oxidation paper, much of it was constructive. The publication of that paper was not a failure of science. Quite the opposite - we learned some valuable lessons about handling this reagent. As far as I can tell the paper was a truthful reporting of their results.
Where this was a failure lies in the way conventional scientific channels handled the matter. There was no mechanism to comment directly on the website where paper was posted. That would have been the logical place for the community to ask questions and have the authors respond. Instead the paper was withdrawn without explanation.
Labels: chemical information, cheminformatics, trust



