CombiUgi and Closing the Open Science Loop
A few weeks ago I asked Rikesh to kick off the CombiUgi project, to create lists of commercially available boc-protected amino acids, aldehydes, primary amines and isonitriles. He is now done and the links to purchase each compound is provided, in addition to the SMILES code.
What we need to do now is generate the list of 90,000 Ugi products resulting from their combination (not including enantiomers and diastereomers and the free amines resulting after boc deprotection). This can probably be done fairly easily with VBA in Excel but if anyone wants to pitch in using their favorite software that would be appreciated.
Although we have not been successful in cyclizing our Ugi products to diketopiperazines, from what we learned, I expect that we should be able to make any of these 90,000 compounds in less than a week (including shipping time).
By indexing these compounds in relevant search engines (I am working with Chemspider to make this happen) as UsefulChem molecules available upon request (and justification) we have an opportunity to close the loop on a practical Open Science project.
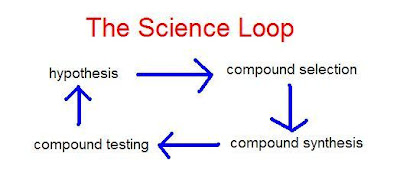 By the loop I mean a complete iteration from hypothesis to deciding which compounds to make to actually making them and getting testing results. These results will confirm or force a modification of the hypothesis and the cycle goes through another iteration hopefully closer to producing a useful outcome (a good drug lead compound for example).
By the loop I mean a complete iteration from hypothesis to deciding which compounds to make to actually making them and getting testing results. These results will confirm or force a modification of the hypothesis and the cycle goes through another iteration hopefully closer to producing a useful outcome (a good drug lead compound for example).I imagine that this loop operates in a lot of research groups. But doing the work under Open Science conditions lets it evolve in new ways. First of all, the direction of progress is determined by the collaborators that elect to participate in the process, not necessarily scientific objectives.
An example of that is our recent shift from the testing of our compounds as anti-malarial agents to testing them as tumor inhibitors simply because Dan Zaharevitz from NCI contacted me and suggested that we submit our compounds.
Right after we started to submit our compounds, Dan left this message:
The folks at Indiana have done a lot of cool stuff that is well worth looking at. One thing they have running in a preliminary form is a service that predicts a
compound's activity in cell lines in the screen. This compound is predicted to be inactive in the cell lines in the prediction. I actually don't think that is a bad result. We probably should put up a place to discuss screens and screening strategy, but essentially a prediction tools such as this summarizes what is known. A compound that is predicted to be inactive, but turns out to be active is much more likely to show you something new and interesting than a compound that is predicted to be active and is active.
So that's the last piece that closes the loop. This web service will make a prediction about activity of the compounds generated by the CombiUgi algorithm and rank them. The flagged compounds will be identifed and synthesized then tested via NCI's assays for tumor cell inhibition.
My groups core expertise is the synthetic component. As far as we are concerned the other 2 processes are black boxes. And for scientists involved in the computation and testing, our synthesis operation is probably a black box. But doing everything in the open, hopefully this will allow other researchers to propose other models and create derivative loops of their own.
We'd love to do the same for the anti-malarial assays but we have not found an established system in place like NCI that will do substrate screening routinely at no cost (except shipping of course).
Is it becoming clearer why I think the scientific process can be automated in novel and useful ways with the progressive adoption of Open Science?




8 Comments:
I am assuming you have a list of R1 groups through to R5 groups? i.e. n1*n2*n3*n4*n5 products? If so the enumeration could probably be done rapidly using macro processing on SMILES - David Jessop has done this here. I have also hacked Markush enumeration into CMLFrag - this means that 3D strucures can be generated although it takes somewhat longer. Please let us know if this is of interest - no absolute promises. Peter Murray-Rust
This comment has been removed by the author.
As PMR pointed out, it's not too difficult to process the SMILES to generate a virtual library.
Andrew Dalke has described a solution using the Python interface to OEChem here.
But it should be equally doable using Openbabel (or even awk for that matter!)
Also, regarding the NCI DTP predictive models that Dan linked to, right now, we only predict for 40 of the 60 cell lines - but we're hoping to get some changes done to get a full panel of 60 predictions
SMILIB is also very useful for this:
http://gecco.org.chemie.uni-frankfurt.de/smilib/index.html
Actually, it really is a matter of quick scripting and you don't even need any cheminformatics libraries :)
I posted a message on the CombiUgi discussion page, but it looks like it messed up the formatting of my Python code.
You can get the code here. it's not the most efficient code and not complete - I couldn't find downloadable SMILES, so I just used the 20 amines and a few of the others (giving 171 compounds).
Could you elaborate on more informatics problems your group may be facing? The more mundane, the better. My own experience as a synthetic organic chemist revealed numerous cases where real chemical information problems went unsolved.
As Rajarshi's response shows, the solutions to many of them are not difficult to implement. Solving these kinds of real, but "unsexy" problems, and making these solutions available to the average chemist, is a big opportunity for those who can recognize it.
Thanks to everyone for their contributions! I've summarized what you have done in a new post.
Rich - I'll try to be as specific as I can in answering your question about what we need in terms of cheminformatics tools:
1) Generate complete SMILES list of Ugi products (as a web service maybe?) based on input from lists of amines, isocyanides, aldehydes and acids. (see complete info on CombiUgi page) Rajarshi has already done this (in Python?) and produced a file of 68,000 compounds but requires that the SMILES of the starting materials position the functional group on the left and currently he has done manually. Code that would automate this process, either as a separate action or integrated with the production of the complete list of Ugi products would be useful.
2) Carry out docking studies from a list of Ugi products (thousands to millions) and malarial enoyl reductase, as done in D-EXP005 with THINK (Sean) and FlexX (Tan). Results from other docking software or other disease targets is welcome.
I've posted these in our ToDo list under the cheminformatics section. Please feel free to update the wiki to comment.
Post a Comment
<< Home